Genetic variant summation
This tool provides access to summary values of DNA mutation analyzed from exome sequencing.
Variant percentages for a single gene for each cell line are calculated using the percent presence of each variant.
If there are multiple variants present, it is presumed that these probabilities are independent,
and that the maximum effect of the variant(s) may go to 100%. The equation used to calculate this probability for
each cell line is: Gene variant percentage per cell = 1 - [ (1 - P1) x (1 - P2) ... (1 - Pn) ] where P1 is the
percentage for variant 1, P2 is the percentage for variant 2, etc. and percentages are taken as fractions (e.g. 25%=0.25).
Variant percentages for multiple genes are simple summations of the single gene values for each cell line.
These percentages can take values greater than 100%. The assumption is that the pathway or functional grouping of
genes will be increasingly perturbed as more of the genes included in it are altered in a functionally meaningful manner.
The tool provides an Excel spreadsheet for each gene that includes:
- a summation of amino acid changing DNA variants (missense, splicesense, frameshift, premature stops,
or nucleotides inserted or deleted in factors of 3 that do not occur in segmental duplications, or map to more than
one genomic location) for up to 150 genes selected by the user
- a summation of amino acid changing variants restricted to those variants that are proposed to be deleterious
and absent from non-cancerous genomes (the 1000 Genomes and EPS5400). Deleterious status is determined by SIFT or
polyphen-2 prediction, or by the presence of a frameshift, splicing, or nonsense change.
|
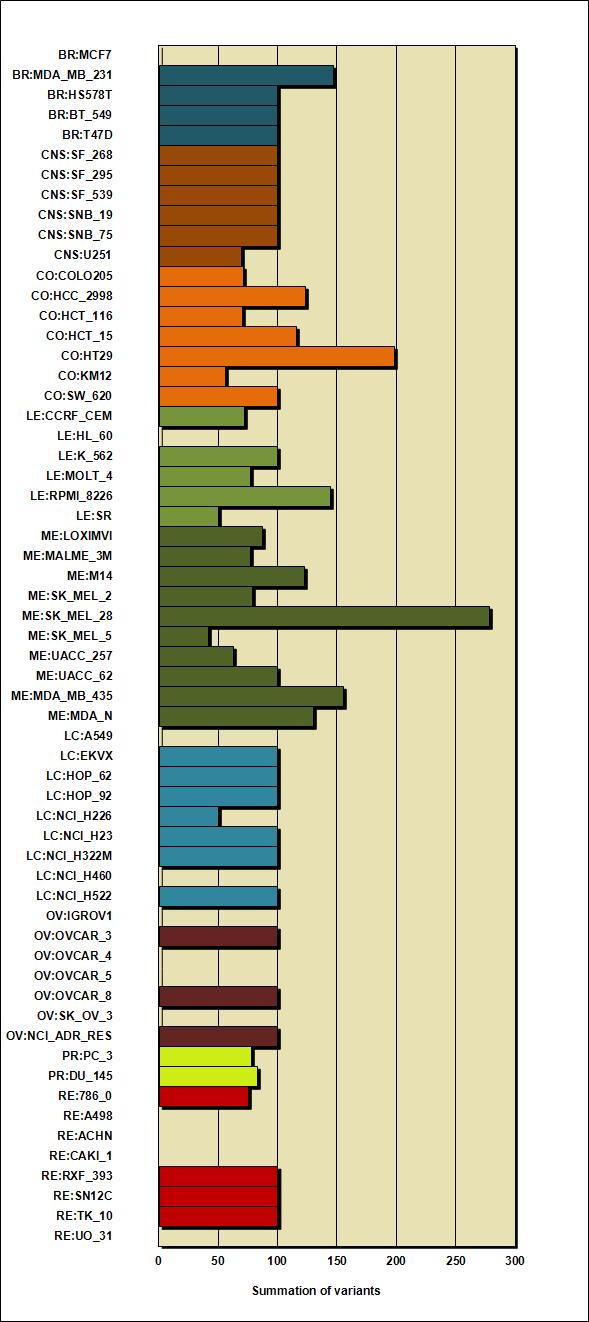
Table 1: Summation of amino acid changing variant(s) |
|
| BR:MCF7 |
0 |
| BR:MDA_MB_231 |
147 |
| BR:HS578T |
100 |
| BR:BT_549 |
100 |
| BR:T47D |
100 |
| CNS:SF_268 |
100 |
| CNS:SF_295 |
100 |
| CNS:SF_539 |
100 |
| CNS:SNB_19 |
100 |
| CNS:SNB_75 |
100 |
| CNS:U251 |
69 |
| CO:COLO205 |
71 |
| CO:HCC_2998 |
123 |
| CO:HCT_116 |
70 |
| CO:HCT_15 |
116 |
| CO:HT29 |
198 |
| CO:KM12 |
56 |
| CO:SW_620 |
100 |
| LE:CCRF_CEM |
72 |
| LE:HL_60 |
0 |
| LE:K_562 |
100 |
| LE:MOLT_4 |
77 |
| LE:RPMI_8226 |
144 |
| LE:SR |
50 |
| ME:LOXIMVI |
87 |
| ME:MALME_3M |
77 |
| ME:M14 |
122 |
| ME:SK_MEL_2 |
79 |
| ME:SK_MEL_28 |
278 |
| ME:SK_MEL_5 |
42 |
| ME:UACC_257 |
63 |
| ME:UACC_62 |
100 |
| ME:MDA_MB_435 |
155 |
| ME:MDA_N |
130 |
| LC:A549 |
0 |
| LC:EKVX |
100 |
| LC:HOP_62 |
100 |
| LC:HOP_92 |
100 |
| LC:NCI_H226 |
50 |
| LC:NCI_H23 |
100 |
| LC:NCI_H322M |
100 |
| LC:NCI_H460 |
0 |
| LC:NCI_H522 |
100 |
| OV:IGROV1 |
0 |
| OV:OVCAR_3 |
100 |
| OV:OVCAR_4 |
0 |
| OV:OVCAR_5 |
0 |
| OV:OVCAR_8 |
100 |
| OV:SK_OV_3 |
0 |
| OV:NCI_ADR_RES |
100 |
| PR:PC_3 |
78 |
| PR:DU_145 |
83 |
| RE:786_0 |
75 |
| RE:A498 |
0 |
| RE:ACHN |
0 |
| RE:CAKI_1 |
0 |
| RE:RXF_393 |
100 |
| RE:SN12C |
100 |
| RE:TK_10 |
100 |
| RE:UO_31 |
0 |
|
|
|
|
