What are the MIM software components?
Several MIM software components have been developed to help developers integrate support for the MIM notation into their software projects. A new specification provides more detailed information regarding the properties of individual MIM elements. Building upon this specification, a computerized format is provided as a standardized method for the storage and exchange of MIM diagrams, and it is accompanied by an application programming interface (API), generated through XMLBeans, to help integrate MIM support into other projects. Additionally, a mechanism is provided to determine if MIM diagrams are valid using Schematron.
What is the MIM notation?
The MIM notation is a graphical notation for biological systems for the representation of networks containing multi-protein complexes, protein modifications, and enzymes that are substrates of other enzymes. This graphical representation makes it possible to view all of the many interactions in which a given molecule may be involved, and it can portray competing interactions, which are common in bioregulatory networks. One aim of the MIM notation is to standardize diagrams of biological systems with a fixed set of glyphs with specific semantics. The specific semantics of the MIM notation can be translated into other standardized formats.
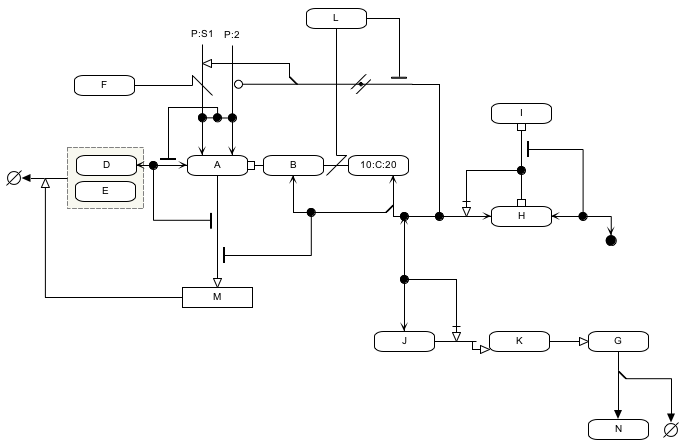
What do MIM diagrams look like?
Here are three sample images:
- the regulation of CAMK and the effect on the phosphorylation of various substrates by the kinase domain of CaMK
- the activation of Chk2 in response to double strand breaks
- an example diagram (shown below) showing all the various elements of the MIM notation, as implemented.
Who are the intended users of the MIM software components?
Programmers wanting to develop software for the MIM notation or to include support for the notation in their software project.
What are the main features of the provided MIM software?
- Provides a standardized schema for the various graphical and non-graphical elements of the MIM notation
- Provides a Java API for parsing, creating, and manipulating instances of MIMML
- Provides a mechanism for the validation of MIMML datasets
- Free and open source

Where can I find the MIMML schema?
Links to the MIM schema are provided below. Documentation for the schema is also provided in a form similar to Javadoc.
Level 1:
Where can I find examples of MIMML datasets?
Example diagrams are provided as MIMML files.What is the MIM API?
The MIM API provides a mechanism for binding MIMML elements to Java objects and provides JavaBeans-style methods such as "getFoo()" and "setFoo()", thereby providing a mechanism for parsing, creating, and manipulating MIMML documents. The MIM API are automatically derived from the MIM XML schemas using Apache Foundation's XMLBeans using version 2.4.0 of XMLBeans. XMLBeans provides a mechanism for binding XML to Java types and provides JavaBeans-style methods such as "getFoo" and "setFoo", thereby providing a mechanism for parsing, creating, and manipulating XML documents.
How do I install the MIM API?
To be used by a Java a program, the mimVis.jar must exist in the classpath a Java program; the .jar file can be downloaded here. Additionally, XMLBeans has two additionally dependencies: jsr173_1.0_api.jar and xbean.jar
Where can I find code samples of using the MIM API?
Sample code is provided on this page in the Code Samples section. Javadoc documentation is a provided as part of the code repository.
Are there bindings for programming languages besides Java?
Currently, we do not maintain any other language bindings. One framework capable of generating bindings for C++ is CodeSynthesis XSD.
Is there a way to validate MIM diagrams?
Schematron is a rule-based validation language for finding patterns in XML trees. Assertions about the presence or absence of these patterns can be used to determine that a document adheres to a given rule set. The Schematron validation schema can be used wherever Extensible Stylesheet Language Family Transformations (XSLT) may be used with XML pipelines and may be used with other standard XML manipulation tools.
How do I know if my diagram is drawn correctly according to the MIM notation?
Validation of MIMML files is done using a Schematron-formatted rule set. The rule set can be used in any environment with an XSLT processor. The rule set is processed into an XSLT stylesheet that is then run against a MIMML file to generate a validation report. The sample command below uses the Xalan XSLT processor
Xalan mimml_validation.sch ./schematron/iso_svrl_for_xslt1.xsl > mimml_validation.xsl; Xalan example.mimml mimml_validation.xsl

Code Samples
The Java code below shows some of the major functionality of the MIM API and below the code are instructions on the compilation of the example.Source Code
The toy example shows how to build a diagram from scratch, how to export the diagram to an XML file, and how to import a valid MIMML file into a diagram object.
package gov.nih.nci.lmp.mim.mimExample;
import java.io.File;
import java.io.FileOutputStream;
import java.io.OutputStream;
import java.util.HashMap;
import org.apache.xmlbeans.XmlOptions;
import gov.nih.nci.lmp.mim.mimVisLevel1.*;
/**
* An class that shows an toy example for the usage of the MIM API.
*/
public class CaCamExample {
/** The MIM-Vis doc. */
public DiagramDocument visDoc;
/**
* Create a DiagramDocument from scratch for the binding of the simple
* physical entity (SPE), Ca++, binding reversibly to another SPE, CaM.
*/
public void createCaCam() {
visDoc = DiagramDocument.Factory.newInstance();
// Create a new diagram to go with the document and set its properties.
DiagramType dia = visDoc.addNewDiagram();
dia.setWidth(84.7);
dia.setHeight(146.3);
// Create a new entity and add it to the diagram.
EntityGlyphType ca = dia.addNewEntityGlyph();
ca.setVisId("b4357");
ca.setCenterX(47.0);
ca.setCenterY(38.0);
ca.setWidth(60.0);
ca.setHeight(20.0);
ca.setColor("000000");
// This shows the way to access enumerated sets from the schema.
ca.setType(EntityGlyphType.Type.SIMPLE_PHYSICAL_ENTITY);
ca.setDisplayName("Ca++");
EntityGlyphType cam = dia.addNewEntityGlyph();
cam.setVisId("a6fc4");
cam.setCenterX(47.0);
cam.setCenterY(123.0);
cam.setWidth(60.0);
cam.setHeight(20.0);
cam.setColor("000000");
cam.setType(EntityGlyphType.Type.SIMPLE_PHYSICAL_ENTITY);
cam.setDisplayName("CaM");
InteractionGlyphType binding = dia.addNewInteractionGlyph();
binding.setVisId("id4f7e9fd7");
binding.setColor("000000");
// Points are used to connect entities to interactions
// and dictate the connection point on entities
InteractionGlyphType.Point pt1 = binding.addNewPoint();
pt1.setX(47.0);
pt1.setY(48.0);
pt1.setArrowHead(ArrowHeadEnumType.NON_COVALENT_REVERSIBLE_BINDING);
pt1.setVisRef("b4357");
//
pt1.setRelX(0.0);
pt1.setRelY(1.0);
InteractionGlyphType.Point pt2 = binding.addNewPoint();
pt2.setX(47.0);
pt2.setY(113.0);
pt2.setArrowHead(ArrowHeadEnumType.NON_COVALENT_REVERSIBLE_BINDING);
pt2.setVisRef("a6fc4");
pt2.setRelX(0.0);
pt2.setRelY(-1.0);
}
/**
* Export file from the DiagramDocument visDoc provided by the MIM API.
*
* @param file
*/
public void exportFile(File file) {
// Set some default export parameters
XmlOptions xmlOptions = new XmlOptions();
try {
OutputStream output = new FileOutputStream(file);
visDoc.save(output, xmlOptions);
output.close();
} catch (Exception e) {
e.printStackTrace();
}
}
/**
* Import file into the DiagramDocument visDoc provided by the MIM API.
*
* @param file
*/
public void importFile(File file) {
try {
visDoc = DiagramDocument.Factory.parse(file);
} catch (Exception e) {
e.printStackTrace();
}
System.out.println("==SUCCESS==");
System.out.println("Number of Entities: "
+ visDoc.getDiagram().sizeOfEntityGlyphArray());
System.out.println("Number of Interactions: "
+ visDoc.getDiagram().sizeOfInteractionGlyphArray());
System.out.println(visDoc.xmlText());
}
/**
* @param args
*/
public static void main(String[] args) {
CaCamExample c = new CaCamExample();
File f = new File(args[1]);
if (args[0].equals("-e")) {
c.createCaCam();
c.exportFile(f);
} else if (args[0].equals("-i")) {
c.importFile(f);
}
}
}
Compilation
Below is the command-line for compiling the example given above; assuming the same directory structure that exists in MIM API code repository. The classpath requires the three .jar files. jsr173_1.0_api.jar and xbean.jar are available from the project SVN repository or can be downloaded as part of XMLBeans. The MIM API, mimVis.jar, is available through the SVN repository in the downloads section. Additionally, an Ant target compile-example is also provided as part of the build.xml in the project SVN repository.javac -classpath lib/jsr173_1.0_api.jar:lib/xbean.jar:mimVis.jar -d build/ src/gov/nih/nci/lmp/mim/mimExample/CaCamExample.java

Where can I download the MIM Pathvisio plugin and how can I get the source code for my own project?
- Download the MIM schema
- Download the MIM API .jar file
- An online code repository for MIM-related projects
- Download the MIMML Schematron rule set
- Download MIMML files: Chk2 and MIM example diagrams
- Tell a friend
- Learn about Schematron
- Learn about XMLBeans
- Report a bug

Authors
Augustin Luna, BS
2005-2007: Postbaccalaureate Intramural Research Training Award Fellow
Clinical Brain Disorders Branch (CBDB)/Genes, Cognition, and Psychosis
Program (GCAP), National Institute of Mental Health (NIMH), National
Institutes of Health (NIH)
2007-Current: Pre-Doctoral Intramural Research Training Award Fellow
Center for Cancer Research (CCR), Laboratory of Molecular Pharmacology (LMP), National Cancer Institute (NCI), National
Institutes of Health (NIH)
BS, Biomedical Engineering, Georgia Institute of Technology
PhD candidate, Bioinformatics, Boston University
Margot Sunshine
SRA
