Gene DNA methylation values
This tool provides access to the DNA methylation data analyzed on the Illumina 450K methylation array.
The tool provides an Excel spreadsheet for each gene that includes:
- the estimated DNA methylation values in tabular and graphical forms (see below)
- the normalized data for those probes that fall within a gene of interest.
The data is queryable by gene or chromosomal location using Query Genomic Data under "DNA: Illumina 450K methylation" (http://discover.nci.nih.gov/cellminer/queryLoad.do).
The data can be downloaded in total at Download Data Sets under "DNA: Illumina 450K methylation" (http://discover.nci.nih.gov/cellminer/loadDownload.do).
A brief platform description is available in Data Set Metadata under "DNA: Illumina 450K methylation" (http://discover.nci.nih.gov/cellminer/datasets.do).
The gene methylation levels are measured as described in ...
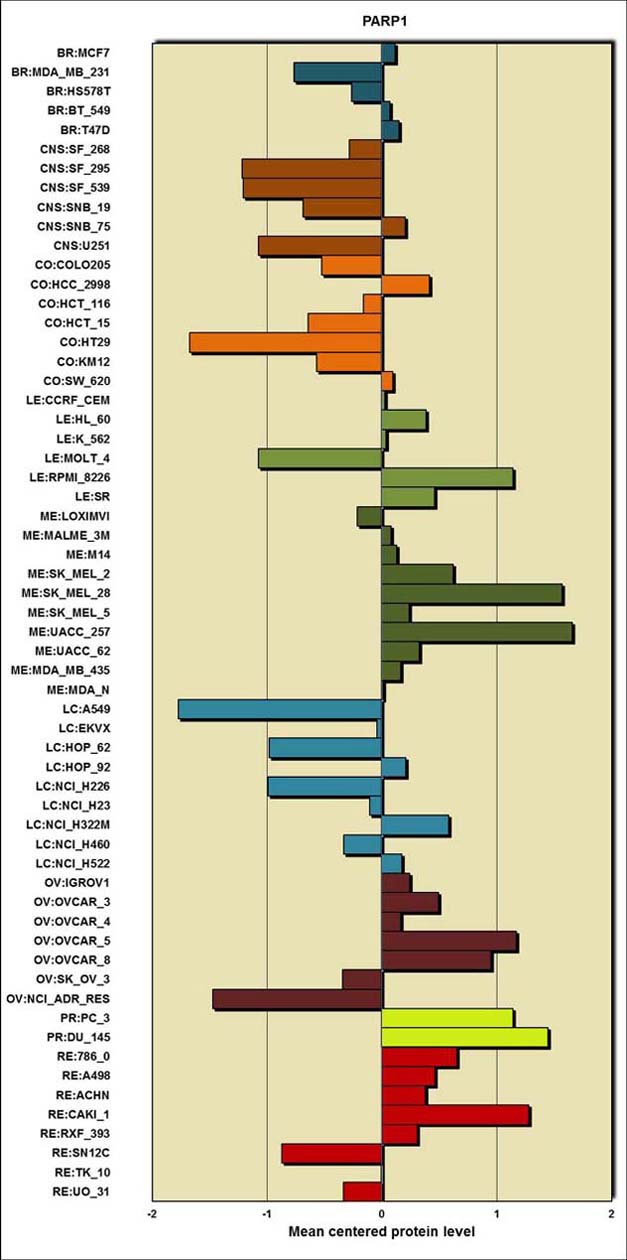
|
| Table 1: Gene methylation levels |
|
| BR:MCF7 |
0.114 |
| BR:MDA_MB_231 |
-0.763 |
| BR:HS578T |
-0.263 |
| BR:BT_549 |
0.073 |
| BR:T47D |
0.151 |
| CNS:SF_268 |
-0.282 |
| CNS:SF_295 |
-1.216 |
| CNS:SF_539 |
-1.208 |
| CNS:SNB_19 |
-0.683 |
| CNS:SNB_75 |
0.204 |
| CNS:U251 |
-1.071 |
| CO:COLO205 |
-0.522 |
| CO:HCC_2998 |
0.415 |
| CO:HCT_116 |
-0.162 |
| CO:HCT_15 |
-0.643 |
| CO:HT29 |
-1.675 |
| CO:KM12 |
-0.565 |
| CO:SW_620 |
0.098 |
| LE:CCRF_CEM |
0.028 |
| LE:HL_60 |
0.385 |
| LE:K_562 |
0.039 |
| LE:MOLT_4 |
-1.073 |
| LE:RPMI_8226 |
1.141 |
| LE:SR |
0.46 |
| ME:LOXIMVI |
-0.212 |
| ME:MALME_3M |
0.082 |
| ME:M14 |
0.129 |
| ME:SK_MEL_2 |
0.62 |
| ME:SK_MEL_28 |
1.57 |
| ME:SK_MEL_5 |
0.24 |
| ME:UACC_257 |
1.656 |
| ME:UACC_62 |
0.327 |
| ME:MDA_MB_435 |
0.1163 |
| ME:MDA_N |
0.013 |
| LC:A549 |
-1.77 |
| LC:EKVX |
-0.041 |
| LC:HOP_62 |
-0.981 |
| LC:HOP_92 |
0.211 |
| LC:NCI_H226 |
-0.988 |
| LC:NCI_H23 |
-0.108 |
| LC:NCI_H322M |
0.583 |
| LC:NCI_H460 |
-0.33 |
| LC:NCI_H522 |
0.174 |
| OV:IGROV1 |
0.241 |
| OV:OVCAR_3 |
0.494 |
| OV:OVCAR_4 |
0.166 |
| OV:OVCAR_5 |
1.174 |
| OV:OVCAR_8 |
0.953 |
| OV:SK_OV_3 |
-0.342 |
| OV:NCI_ADR_RES |
-1.469 |
| PR:PC_3 |
1.14 |
| PR:DU_145 |
1.449 |
| RE:786_0 |
0.652 |
| RE:A498 |
0.461 |
| RE:ACHN |
0.38 |
| RE:CAKI_1 |
1.279 |
| RE:RXF_393 |
0.307 |
| RE:SN12C |
-0.869 |
| RE:TK_10 |
-0.006 |
| RE:UO_31 |
-0.335 |
|
|
|
|
